

Assignees.I have Illumina paired-end reads for E. 18 (r982:295) Usage: samtools Command: view SAMBAM conversion sort sort alignment file mpileup multi-way pileup depth compute the depth faidx index/extract FASTA tview text alignment viewer index index alignment idxstats BAM index stats (r595 or later) fixmate fix mate information flagstat simple SAMTOOLS - COVERAGE This application computes the depth at each position or region andproduces a histogram or table of coverage per chromosome from an input BAM file.
BAM FILE FORMAT CODE
Here's some code to use Samtools to extract the sequencing coverage depth from a BAM file for multiple regions of interest as specified in a BED file.To extract the sequencing coverage for each position and subsequently calculate the median coverage for each sample, Samtools depth v1.at least 5X) 20X considered high-coverage
BAM FILE FORMAT FULL
Show only ASCII characters in histogram using colon and fullstop for full and half height characters.
BAM FILE FORMAT WINDOWS
For example, bedtools coverage can compute the coverage of sequence alignments (file B) across 1 kilobase (arbitrary) windows (file A) tiling a genome of interest. Note: Brent Pedersen wrote mosdepth (publication here) which should be faster than samtools depth especially for large mappings (such as genome).
BAM FILE FORMAT HOW TO
Finally, we wrap up this chapter with a description of the program bedtools, and how to use it to investigate the depth of reads aligning to different portions of the reference genome. For more information see SAMtools documentation. d,-min-depth INT Specifies the minimum base depth to consider a reference position to be covered, for purposes of the FRPERC and CRPERC sections.

daviesrob mentioned this issue 13 days ago. You also don’t need to store the coverage into an intermediate file, thus reducing IO cost: sum=$(samtools depth "$1" | awk '' aligned. 01, meaning depths within +/- 1% of a mid point will be Samtools’s mpileup (formerly pileup) computes depth of coverage (eg.
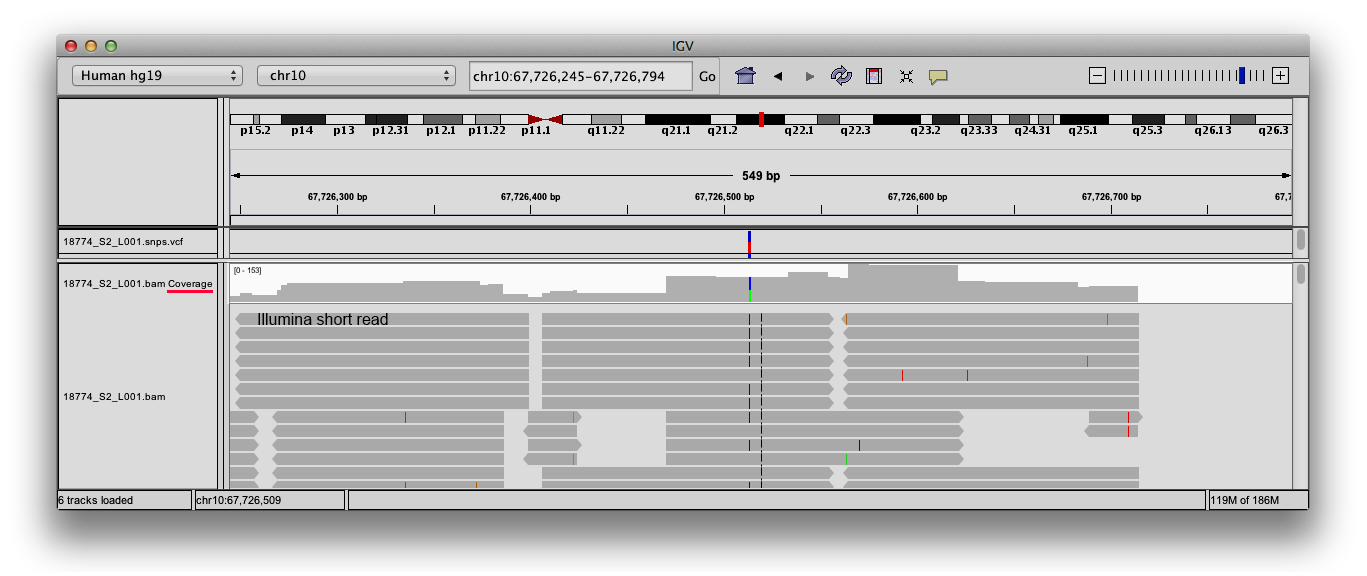
The depth command in the current version of samtools produces three tab delimited columns. 01, meaning depths within +/- 1% of a mid point will be

The first is the name of the contig or chromosome, the second is the position, and the third is the number of reads aligned at that position. txt gives coverage data for each base in the region that is in the BAM file. pl with the varFilter -D100 option, which filters out SNPs that had read depth higher than 100 (we don’t want to trust SNPs at sites with super high coverage, because they might be bam | grep "contig_youwant_to_count" | gzip > coverage The input BAM files hast to samtools coverage -r chr1:1M-12M input. Samtools depth of coverage bam -counts >.


 0 kommentar(er)
0 kommentar(er)
